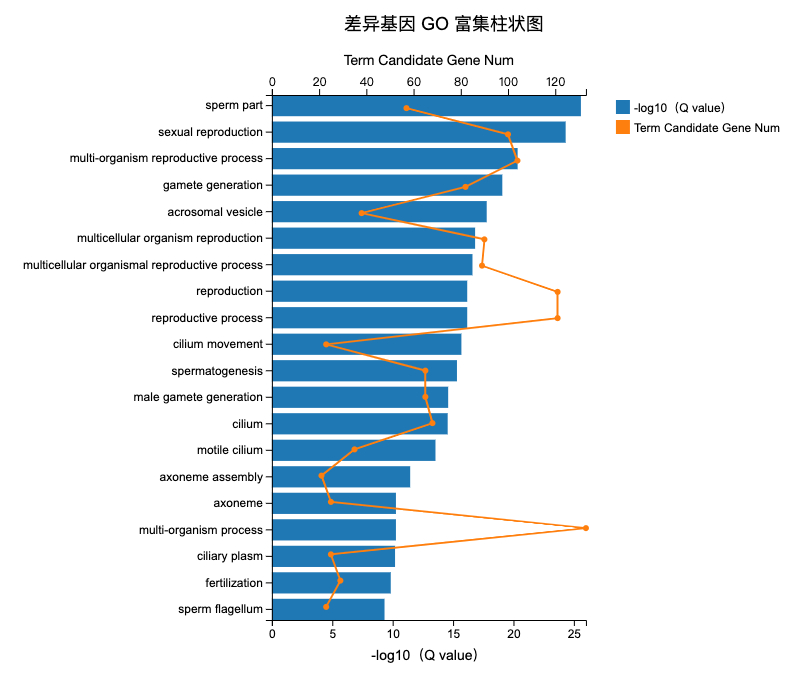
大神请问上图怎么做的?求代码!
以下回答参考 皆我百晓生、券券喵儿 等免费微信小程序相关内容作答,并由本人整理回复。
要使用R语言绘制这些双轴柱状图,你需要先安装并加载ggplot2库。然后,你可以根据你的数据创建一个数据框,并利用ggplot()函数来绘制图形。
以下是一个基本的示例,展示了如何创建这两个图表:
# 假设你已经有了以下的数据
data <- data.frame(
Term = c("sperm part", "sexual reproduction", "multi-organism reproductive process",
"gamete generation", "acrosomal vesicle", "multicellular organism reproduction",
"reproduction", "reproductive process", "cilium movement", "spermatogenesis",
"male gamete generation", "cilium", "motile cilium", "axoneme assembly", "axoneme",
"multi-organism process", "ciliary plasm", "fertilization", "sperm flagellum"),
CandidateGeneNum = c(20, 40, 60, 80, 100, 120),
Qvalue = c(-log10(0.05), -log10(0.05), -log10(0.05), -log10(0.05), -log10(0.05), -log10(0.05)),
GO = c("sexual reproduction", "sexual reproduction", "sexual reproduction", "sexual reproduction", "sexual reproduction", "sexual reproduction", "reproduction", "reproduction", "reproduction", "reproduction", "reproduction", "reproduction", "reproduction", "reproduction", "reproduction", "reproduction", "reproduction", "reproduction", "reproduction", "reproduction", "reproduction", "reproduction", "reproduction", "reproduction", "reproduction", "reproduction", "reproduction")
)
# 创建数据框
df <- data.frame(
Term = paste(data$Term, collapse = ""),
CandidateGeneNum = data$CandidateGeneNum,
Qvalue = data$Qvalue,
GO = data$GO
)
# 使用 ggplot2 绘制第一个图表
ggplot(df, aes(x=Term, y=CandidateGeneNum)) +
geom_bar(stat="identity") +
xlab("") + ylab("") +
theme_minimal() +
scale_y_continuous(labels=paste0("n=", round(10^x))) +
labs(title = "Term and Number of Candidate Genes with Significant GO Terms")
# 使用 ggplot2 绘制第二个图表
ggplot(df, aes(x=Term, y=Qvalue)) +
geom_bar(stat="identity") +
xlab("") + ylab("") +
theme_minimal() +
scale_x_discrete(NULL, labels=c("-log10(0.05)", "-log10(0.05)", "-log10(0.05)", "-log10(0.05)", "-log10(0.05)", "-log10(0.05)")) +
labs(title = "Term and Q-value of GO Terms")
这个代码将生成两个图表,分别显示了差异基因的GO富集情况和差异基因的Q值。请注意,你可能需要根据你的具体数据调整这些参数以获得最佳效果。