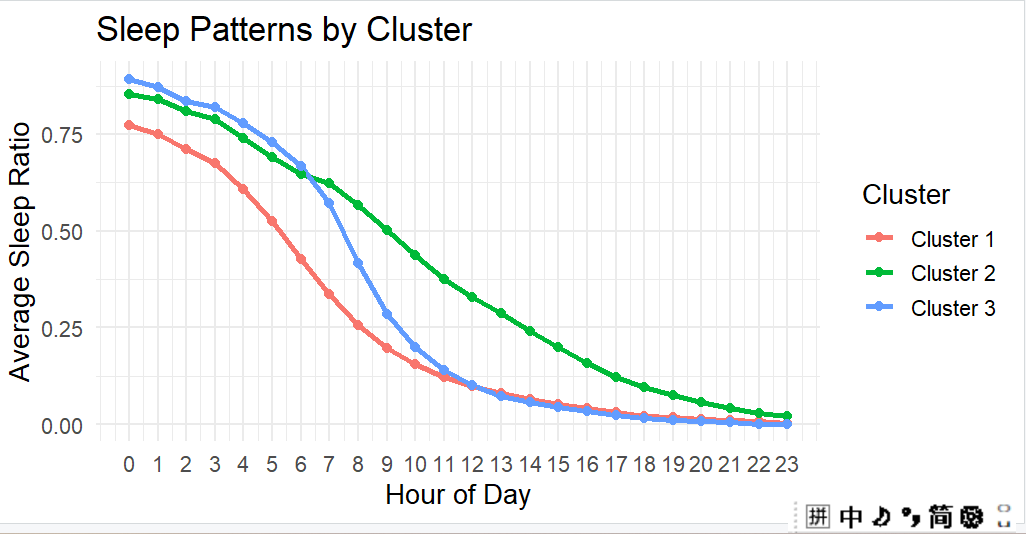
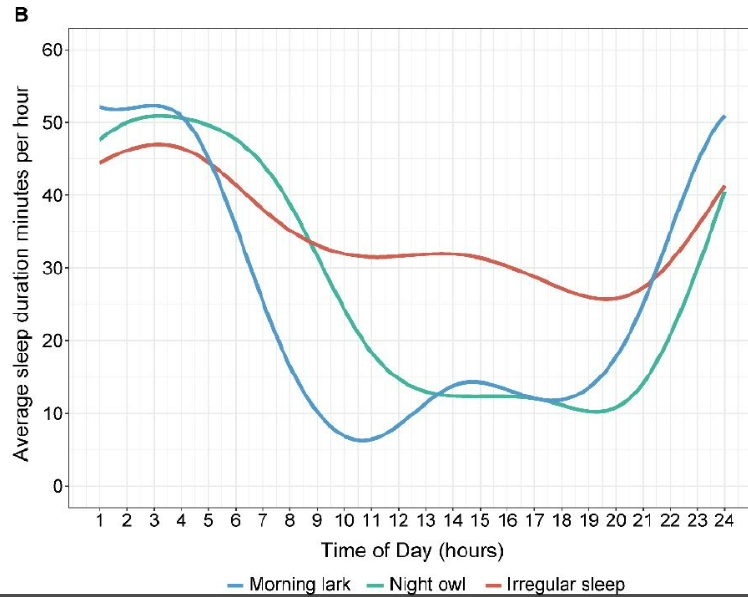
我们想要利用加速度计数据进行睡眠模式的聚类。但是图一是我们自己聚类的(sleep patterns by cluster),图二是文献中符合正常的聚类图。原始文献中提到,他们使用的方法是使用已经处理好的加速度计分钟级别数据,对于睡眠模式,我们计算了每小时累积的睡眠分钟数的比例作为自变量,采用欧几里得距离为标准进行KM聚类分析,请问,我们的处理过程哪里出现了问题呢?应如何修改呢?以下是我们的处理代码:
#下载2011周期数据----
#下载数据----
library(haven) # 读取 XPT
library(data.table)
library(dplyr)
library(lubridate)
# ---- Step 1. 分批读取(节省内存) ----
# 先载入数据
pax_path <- "PAXMIN_G.xpt"
pax <- as.data.table(read_xpt(pax_path))
# 保留关键变量
pax <- pax[, .(SEQN, PAXDAYM, PAXDAYWM, PAXMTSM, PAXQFM, PAXPREDM)]
# PAXPREDM: 1=wake, 2=sleep, 3=nonwear, 4=unknown(详见 codebook)
# 检查基本情况
pax[, .N, by = PAXPREDM]
head(pax)
#检查每个受试者的天数
pax[, .(min_day = min(as.numeric(PAXDAYM)),
max_day = max(as.numeric(PAXDAYM)),
n = .N),
by = SEQN][1:10]
# Step 2: 转换状态编码为 factor,方便可视化
pax[, state := factor(PAXPREDM,
levels = c("1", "2", "3", "4"),
labels = c("Wake", "Sleep", "Nonwear", "Unknown"))]
#排序
pax <- pax[order(SEQN, PAXDAYM, PAXMTSM)]
# ---- Step 3. 数据清理 ----
# MIMS 值(PAXMTSM)用于分析但不是时间索引,因此设 NA 不影响时间
pax[, PAXMTSM := fifelse(PAXMTSM == -0.01, NA_real_, PAXMTSM)]
pax[state == "Nonwear", PAXMTSM := NA_real_]
pax[PAXQFM > 0, PAXMTSM := NA_real_]
# ---- Step 4. 有效天判定(使用 PAXMTSM 是否缺失) ----
valid_days <- pax[, .(
wear_minutes = sum(!is.na(PAXMTSM))
), by = .(SEQN, PAXDAYM)]
valid_days[, valid_day := wear_minutes >= 960]
valid_subjects <- valid_days[, .(
valid_day_count = sum(valid_day)
), by = SEQN]
valid_subjects <- valid_subjects[valid_day_count >= 4]
pax <- pax[SEQN %in% valid_subjects$SEQN]
cat("有效受试者数量:", nrow(valid_subjects), "\n")
# ============================
# 正确生成 minute_of_day 和 hour
# ============================
pax_valid <- copy(pax)
# 正确生成分钟序号 (每个受试者、每天从 0 到 1439)
pax_valid[, minute_of_day := seq_len(.N) - 1, by = .(SEQN, PAXDAYM)]
# 小时 = 0 到 23,不会有 NA
pax_valid[, hour := minute_of_day %/% 60]
# Sleep flag
pax_valid[, sleep_flag := fifelse(state == "Sleep", 1, 0)]
# 按 SEQN × 日期 × 小时 聚合
hourly_sleep <- pax_valid[, .(
sleep_minutes = sum(sleep_flag, na.rm = TRUE),
total_minutes = .N
), by = .(SEQN, PAXDAYM, hour)]
hourly_sleep[, sleep_ratio := sleep_minutes / total_minutes]
# 每个受试者跨天平均
subject_hour_avg <- hourly_sleep[, .(
avg_sleep_ratio = mean(sleep_ratio, na.rm = TRUE)
), by = .(SEQN, hour)]
summary(subject_hour_avg)
saveRDS(subject_hour_avg, file = "subject_hour_avg.rds")
# 下载2013-2014周期数据 ----
library(haven)
library(data.table)
library(dplyr)
library(lubridate)
# ---- Step 1. 分批读取 ----
pax_I_path <- "PAXMIN_H.xpt"
pax_I <- as.data.table(read_xpt(pax_I_path))
# 保留变量
pax_I <- pax_I[, .(SEQN, PAXDAYM, PAXDAYWM, PAXMTSM, PAXQFM, PAXPREDM)]
# 检查
pax_I[, .N, by = PAXPREDM]
head(pax_I)
pax_I[, .(min_day = min(as.numeric(PAXDAYM)),
max_day = max(as.numeric(PAXDAYM)),
n = .N),
by = SEQN][1:10]
# Step 2: 状态编码
pax_I[, state := factor(PAXPREDM,
levels = c("1", "2", "3", "4"),
labels = c("Wake", "Sleep", "Nonwear", "Unknown"))]
# 排序
pax_I <- pax_I[order(SEQN, PAXDAYM, PAXMTSM)]
# ---- Step 3. 数据清理 ----
pax_I[, PAXMTSM := fifelse(PAXMTSM == -0.01, NA_real_, PAXMTSM)]
pax_I[state == "Nonwear", PAXMTSM := NA_real_]
pax_I[PAXQFM > 0, PAXMTSM := NA_real_]
# ---- Step 4. 有效天判定 ----
valid_days_I <- pax_I[, .(
wear_minutes = sum(!is.na(PAXMTSM))
), by = .(SEQN, PAXDAYM)]
valid_days_I[, valid_day := wear_minutes >= 960]
valid_subjects_I <- valid_days_I[, .(
valid_day_count = sum(valid_day)
), by = SEQN]
valid_subjects_I <- valid_subjects_I[valid_day_count >= 4]
pax_I <- pax_I[SEQN %in% valid_subjects_I$SEQN]
cat("有效受试者数量(2013–2014):", nrow(valid_subjects_I), "\n")
# ---- 正确生成 minute_of_day 与 hour ----
pax_I_valid <- copy(pax_I)
pax_I_valid[, minute_of_day := seq_len(.N) - 1, by = .(SEQN, PAXDAYM)]
pax_I_valid[, hour := minute_of_day %/% 60]
pax_I_valid[, sleep_flag := fifelse(state == "Sleep", 1, 0)]
# ---- 小时聚合 ----
hourly_sleep_I <- pax_I_valid[, .(
sleep_minutes = sum(sleep_flag, na.rm = TRUE),
total_minutes = .N
), by = .(SEQN, PAXDAYM, hour)]
hourly_sleep_I[, sleep_ratio := sleep_minutes / total_minutes]
# ---- 每受试者跨天平均 ----
subject_hour_avg_I <- hourly_sleep_I[, .(
avg_sleep_ratio = mean(sleep_ratio, na.rm = TRUE)
), by = .(SEQN, hour)]
summary(subject_hour_avg_I)
#保存数据----
saveRDS(subject_hour_avg_I, file = "subject_hour_avg_I.rds")
#加载数据----
subject_hour_avg_I<- readRDS("subject_hour_avg_I.rds")
subject_hour_avg <- readRDS("subject_hour_avg.rds")
#合并数据----
library(data.table)
# 读入两个 RDS 数据
dt_A <- readRDS("subject_hour_avg.rds") # 2011–2012
dt_B <- readRDS("subject_hour_avg_I.rds") # 2013–2014
summary(dt_A)
summary(dt_B)
# 合并
dt_all <- rbindlist(list(dt_A, dt_B))
# 展开为 24 小时特征(每个受试者一行 × 24 列)
dt_wide <- dcast(dt_all, SEQN ~ hour, value.var = "avg_sleep_ratio")
summary(dt_wide)
# 去掉 SEQN,只保留数据矩阵
X <- as.matrix(dt_wide[, -1])
rownames(X) <- dt_wide$SEQN
# 检查
dim(X)
head(X[,1:5])
#进行KM聚类分析----
## K-Means for NHANES sleep data
## 改写自你提供的算法
k = 3 # 自定义聚类数
maxIter = 30
epsilon = 0.001
## 计算点到所有质心的距离
centroidDists <- function(X, c) {
dists = apply(X, 1, function(point)
sapply(1:nrow(c), function(dim)
dist(rbind(point, c[dim, ]))))
return(t(dists))
}
## 分配每个点所属的聚类
assignClusters <- function(X, c) {
cDists = centroidDists(X, c)
clusters = apply(cDists, 1, which.min)
return(clusters)
}
## 绘图:如果维度>2,则使用 pairs()
#plotData <- function(X, title, centroids = NULL, clusters = NULL) {
if (ncol(X) > 2) {
if (!is.null(centroids) & !is.null(clusters)) {
pairsCol = append(clusters + 1,
rep(1, nrow(centroids)))
pairs(rbind(X, centroids), main = title, col = pairsCol)
} else {
pairs(X, main = title)
}
} else {
plot(X, col = clusters + 1, main = title)
if (!is.null(centroids))
points(centroids, col = "black", cex = 3)
}
}
## K-means 主程序
cat("数据维度:", dim(X), "\n")
n = nrow(X)
d = ncol(X)
# 随机选择初始质心
ids = sample(1:n, k)
centroids = X[ids, ]
clusters = rep(0, n)
for(iter in 1:maxIter) {
cat("迭代:", iter, "\n")
# 1. 分配聚类
clusters = assignClusters(X, centroids)
# 画图(24维数据会使用 pairs)
#plotData(X, paste("Iteration", iter), centroids, clusters)
# 2. 更新质心
newCentroids = t(sapply(1:k, function(c)
colMeans(X[clusters == c, , drop = FALSE])
))
# 判断是否收敛
delta = sum((newCentroids - centroids)^2)
cat("Delta:", delta, "\n")
if (delta < epsilon) {
cat("已收敛。\n")
break
}
centroids = newCentroids
}
## 最终聚类图
#plotData(X, "Final Clustering", centroids, clusters)
## 输出结果
cat("\n聚类结束。\n\n")
for (c in 1:nrow(centroids)) {
cat("Cluster ", c, ":\n", sep = "")
cat("\tCentroid:", paste(round(centroids[c, ],3), collapse = ", "), "\n")
cat("\tSize:", sum(clusters == c), "\n")
}
cat("\nIteration Count:", iter, "\n")
cat("Final Delta:", delta, "\n")
# 保存聚类编号
cluster_results <- data.table(SEQN = rownames(X), cluster = clusters)
saveRDS(cluster_results, "sleep_clusters.rds")
readRDS("sleep_clusters.rds")
library(ggplot2)
library(tidyr)
library(dplyr)
# 构建数据框
hours <- 0:23
cluster_centroids <- data.frame(
hour = rep(hours, 3),
avg_sleep_ratio = c(
c(0.773, 0.751, 0.712, 0.674, 0.607, 0.524, 0.427, 0.337, 0.255, 0.196, 0.155, 0.122, 0.099, 0.08, 0.065, 0.053, 0.042, 0.031, 0.022, 0.018, 0.014, 0.01, 0.007, 0.004), # Cluster 1
c(0.853, 0.841, 0.81, 0.789, 0.739, 0.69, 0.646, 0.623, 0.567, 0.502, 0.436, 0.374, 0.329, 0.287, 0.242, 0.199, 0.159, 0.123, 0.096, 0.075, 0.057, 0.041, 0.029, 0.021), # Cluster 2
c(0.893, 0.872, 0.836, 0.82, 0.779, 0.73, 0.667, 0.571, 0.417, 0.286, 0.199, 0.14, 0.102, 0.073, 0.056, 0.044, 0.034, 0.023, 0.015, 0.012, 0.009, 0.005, 0.001, 0.001) # Cluster 3
),
cluster = rep(c("Cluster 1", "Cluster 2", "Cluster 3"), each = 24)
)
# 绘图
ggplot(cluster_centroids, aes(x = hour, y = avg_sleep_ratio, color = cluster)) +
geom_line(size = 1.2) +
geom_point(size = 2) +
scale_x_continuous(breaks = 0:23) +
labs(title = "Sleep Patterns by Cluster",
x = "Hour of Day",
y = "Average Sleep Ratio",
color = "Cluster") +
theme_minimal(base_size = 14)