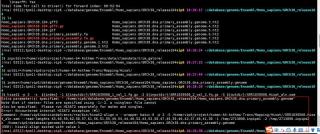
下面为上图的红圈error信息:
Extra parameter(s) specified: "/home/vip6/database/genome/Ensembl/Homo_sapiens/GRCh38_release104/Homo_sapiens.GRCh38_release104.genome"
Note that if <mates> files are specified using -1/-2, a <singles> file cannot
also be specified. Please run HISAT2 separately for mates and singles.
Error: Encountered internal HISAT2 exception (#1)
Command: /home/vip6/miniconda3/envs/rna/bin/hisat2-align-s --wrapper basic-0 -p 3 -S /home/vip6/project/human-64-Asthma-Trans/Mapping/Hisat//SRR1039508.Hisat_aln.sam --read-lengths 63,60,59,62,58,57,61,56,51,55,49,54,44,40,52,43,53,50,47,45,39,46,42,37,48,41,38 -1 /tmp/2597247.inpipe1 -2 /tmp/2597247.inpipe2 ?-x ?/home/vip6/database/genome/Ensembl/Homo_sapiens/GRCh38_release104/Homo_sapiens.GRCh38_release104.genome
(ERR): hisat2-align exited with value 1