# Load required modules
import pandas as pd
import scipy.spatial
import scipy.cluster
import numpy as np
import json
import matplotlib.pyplot as plt
from functools import reduce
# Example data: gene expression
geneExp = {'genes' : ['a', 'b', 'c', 'd', 'e', 'f', 'g', 'h', 'i', 'j'],
'exp1': [-2.2, 5.6, 0.9, -0.23, -3, 0.1, 1.0, 3.0, 1.2, 1.3],
'exp2': [5.4, -0.5, 2.33, 3.1, 4.1, -3.2, -1.0, -1.2, -1.3, -1.1]
}
df = pd.DataFrame( geneExp )
# Determine distances (default is Euclidean)
dataMatrix = np.array( df[['exp1', 'exp2']] )
distMat = scipy.spatial.distance.pdist( dataMatrix )
# Cluster hierarchicaly using scipy
clusters = scipy.cluster.hierarchy.linkage(distMat, method='single')
T = scipy.cluster.hierarchy.to_tree( clusters , rd=False )
# Create dictionary for labeling nodes by their IDs
labels = list(df.genes)
id2name = dict(enumerate(labels))
# Draw dendrogram using matplotlib to scipy-dendrogram.pdf
scipy.cluster.hierarchy.dendrogram(clusters, labels=labels, orientation='right')
plt.savefig("scipy-dendrogram.png")
# Create a nested dictionary from the ClusterNode's returned by SciPy
def add_node(node, parent ):
# First create the new node and append it to its parent's children
newNode = dict( node_id=node.id, children=[] )
parent["children"].append( newNode )
# Recursively add the current node's children
if node.left: add_node( node.left, newNode )
if node.right: add_node( node.right, newNode )
# Initialize nested dictionary for d3, then recursively iterate through tree
d3Dendro = dict(children=[], name="Root1")
add_node( T, d3Dendro )
根据上述代码及demo数据,可获得系统发育树及包含节点信息的字典d3Dendro如下:
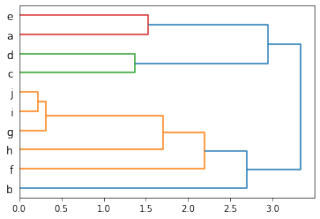
>>> d3Dendro
{'children': [{'children': [{'children': [{'children': [], 'name': 'b'},
{'children': [{'children': [], 'name': 'f'},
{'children': [{'children': [], 'name': 'h'},
{'children': [{'children': [], 'name': 'g'},
{'children': [{'children': [], 'name': 'i'},
{'children': [], 'name': 'j'}],
'name': 'i,j'}],
'name': 'g,i,j'}],
'name': 'g,h,i,j'}],
'name': 'f,g,h,i,j'}],
'name': 'b,f,g,h,i,j'},
{'children': [{'children': [{'children': [], 'name': 'c'},
{'children': [], 'name': 'd'}],
'name': 'c,d'},
{'children': [{'children': [], 'name': 'a'},
{'children': [], 'name': 'e'}],
'name': 'a,e'}],
'name': 'a,c,d,e'}],
'name': 'a,b,c,d,e,f,g,h,i,j'}],
'name': 'Root1'}
我想请问如何根据一段python脚本,自动获取系统发育树的每个节点所对应的两组样本的名称的列表?
对于上述demo数据,目标获取的结果应该是:
[ 'a,c,d,e' , 'b,f,g,h,i,j' ], ['a,e', 'c,d'], ['a', 'e'], ['c', 'd'] ,['b', 'f,g,h,i,j' ], ['f', 'g,h,i,j' ], ['h', 'g,i,j' ], ['g', 'i,j'], ['i', 'j' ]
