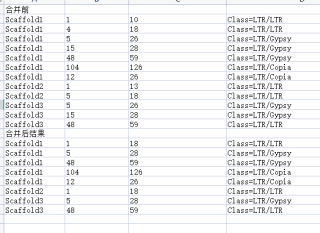
如下表所示,将重复序列进行合并,如果两种重复序列在坐标上是有重合,则将两个重复序列合并成一个,输出。
请写清楚完整代码
我参考网上的写了一个但是发现无法统计染色体结果
use strict;
use warnings;
my @name_array ;
my $hash;
open (FILE, "trf.gff");
while(<FILE>){
my ($scaff, $a, $b, $min, $max, $c, $d, $e, $f, $g, $class, $h, $i, $j) = split(/\t|;/); #是对的
unless (grep $_->[8] eq $class ,@name_array){
push @name_array, [$scaff, $a, $b, $c, $d, $e, $f, $g, $class, $h, $i, $j];
} ;
($min,$max) = ($max,$min) if $min > $max ;
if($hash->{$class}){
sort_matrix($min,$max,$hash->{$class});
}
else{
push @{$hash->{$class}},[$min,$max];
}
}
###print sort result
for my $item( @name_array ){
my $class = $item->[8];
for (0..$#{$hash->{$class}}){
print $item->[0]," ",$item->[1]," ",$item->[2]," ",$hash->{$class}->[$_][0]," ",$hash->{$class}->[$_][1]," ",$item->[3]," ",$item->[4]," ",$item->[5]," ",$item->[6]," ",$item->[7]," ",$class,"_E",($_+1)," ",$item->[9]," ",$item->[10]," ",$item->[11],"\n";
}
}
sub sort_matrix{
my ($cur_min,$cur_max,$matrix) = @_;
# record index
my @splice_index;
for (0..$#$matrix){
my $item_min = $matrix->[$_]->[0];
my $item_max = $matrix->[$_]->[1];
next if $cur_max < $item_min or $cur_min > $item_max;
return if $cur_min >= $item_min and $cur_max <= $item_max;
$cur_min = $cur_min < $item_min ? $cur_min : $item_min;
$cur_max = $cur_max < $item_max ? $item_max : $cur_max;
push @splice_index, $_;
}
push @$matrix,[$cur_min,$cur_max];
#print "@splice_index\n";
for (0..$#splice_index){
splice(@$matrix,$splice_index[$_]-$_,1);##This is trick
}
#print $cur_min,":",$cur_max,"\n";
}